Towards Characterizing Genomics Workload Performance at Scale
Leveraging Chameleon's Bare Metal Resources to Benchmark Genomics Workflows
- Aug. 26, 2024 by
- Martin Putra
Genomics data analysis has potential to empower precision medicine by accelerating the generation of insights from large and diverse amounts of samples. For example, the analysis results can help physicians to predict the existence of cancer at an early stage, and also tailor treatment based on a patient's genetic information. Oncologists and public health specialists working with population-wide genomics data can infer the most common mutation among people in a certain geographic area, and thus guide the design of healthcare systems and policy for that area.
All these efforts benefit from the large amounts of samples, yet from a computational perspective, such analysis is very challenging at least for two reasons: 1.) the massive size of input data, and 2.) the diverse characteristics of genomics applications. This leads to many interesting problems in subareas of workload characterization, performance modeling, scheduling, and reliability to name a few.
Research Overview
One of the problems we are working on right now is the performance characterization of genomics workflows. Genomics workflows are often composed of independent applications with diverse characteristics: they can be I/O bound, compute-bound, memory-bound, or a mix of all. An intimate understanding of such characteristics, especially when the workflows are executed at large scale, is crucial to design effective scheduling & resource management techniques.
To this end, we attempt to build a scalable benchmarking tool that is representative of the scale of a real production system. The underlying intuition is that most of the problems we face happen due to scale, thus all insights we use to solve the problems should also be based on data collected at similar scale, lest our solutions might be misguided. We have built a prototype which enables us to run a genomics workflow using open-source inputs and capture the runtime resource utilization (i.e., CPU, memory, and I/O) of each individual application. Below we show the CPU utilization pattern of 9 longest applications used by a popular genomics workflow.
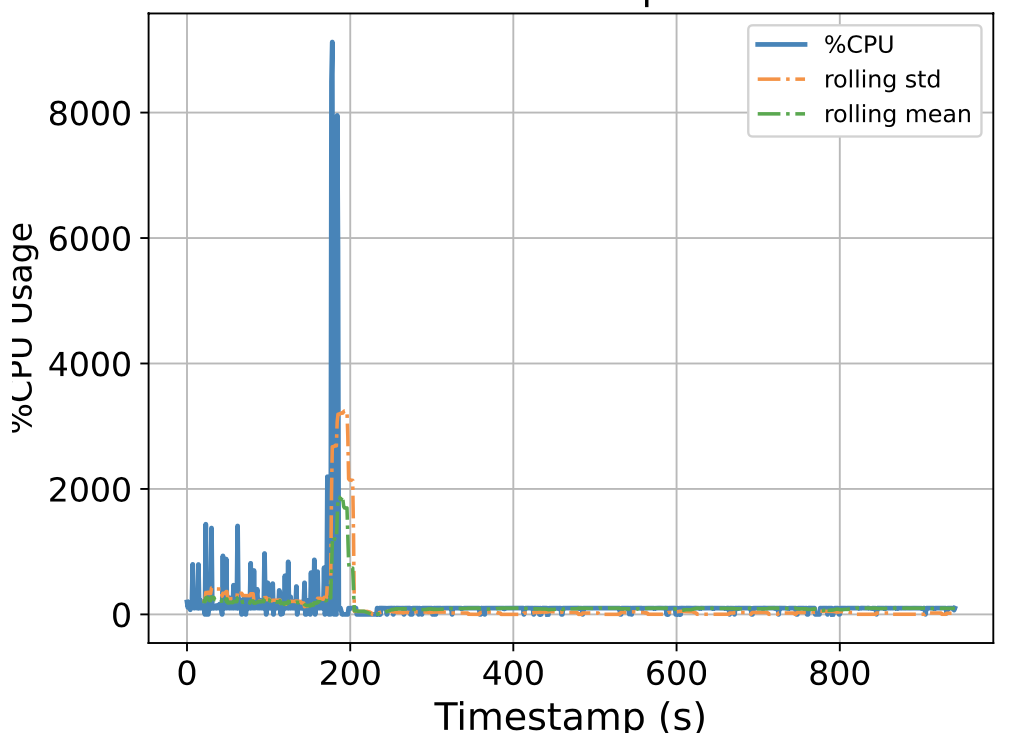
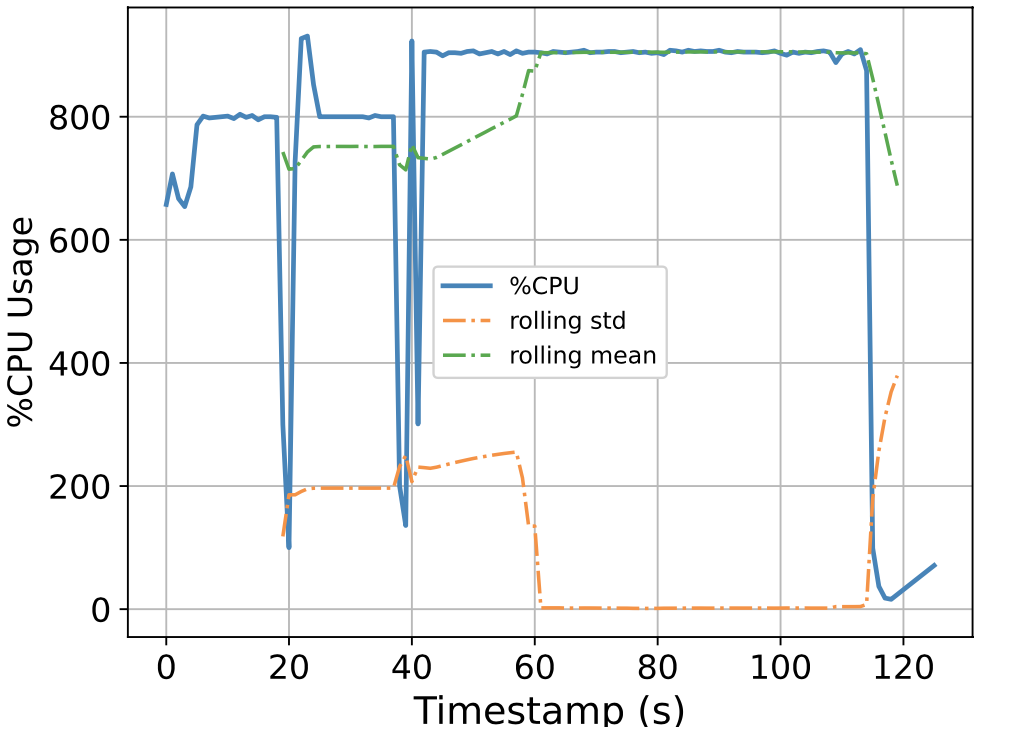
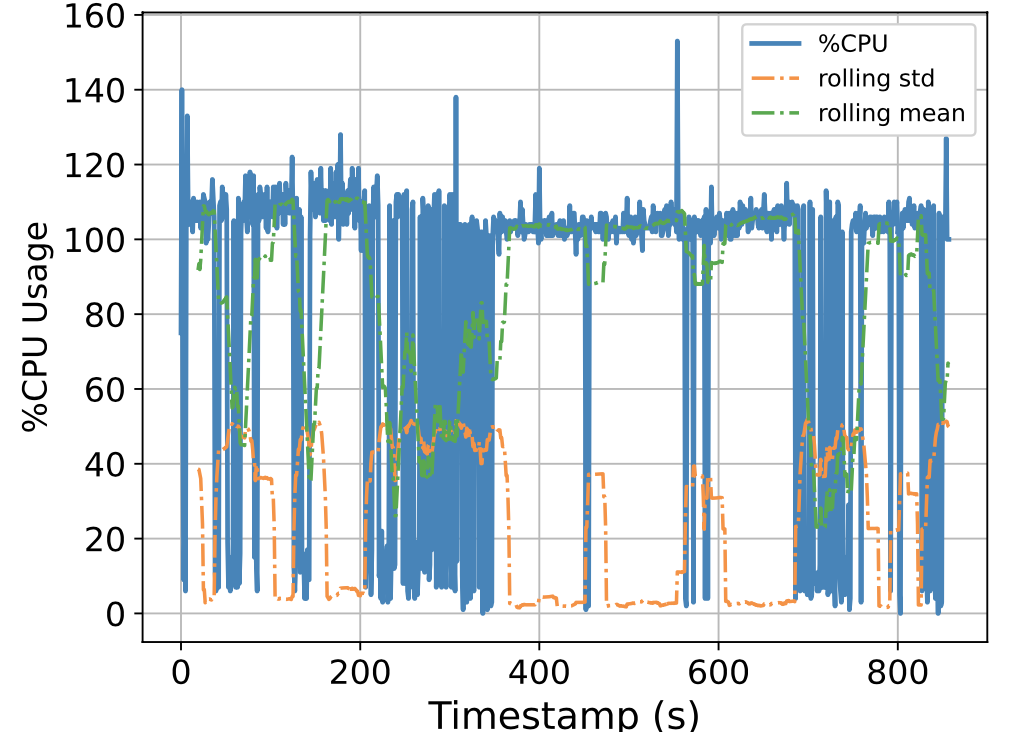
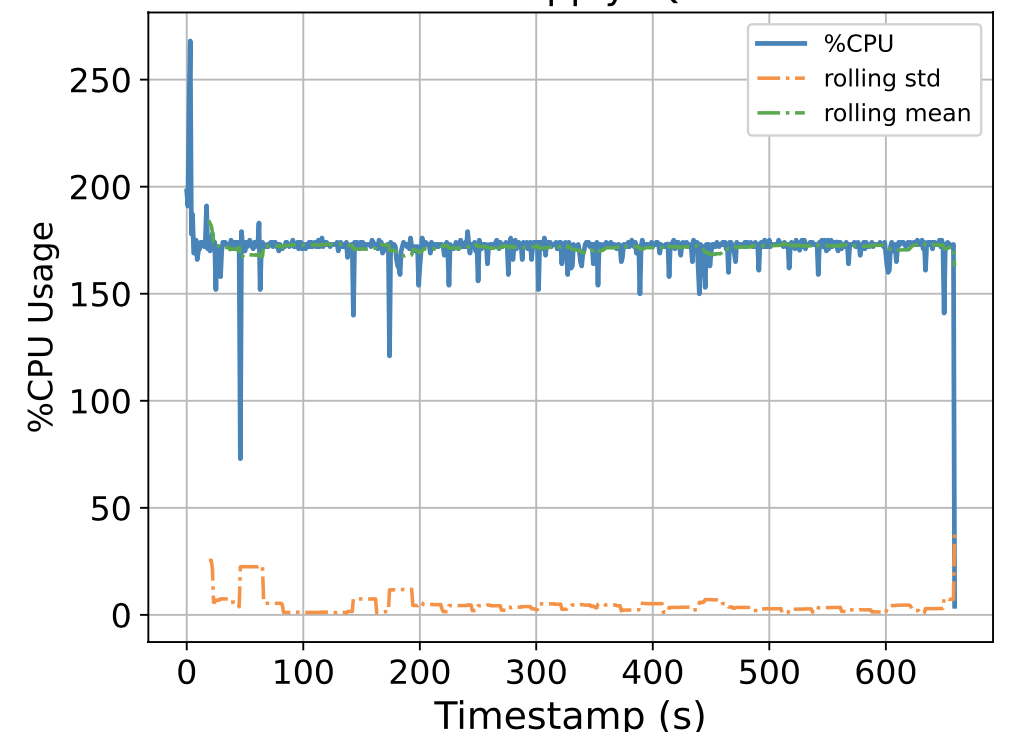
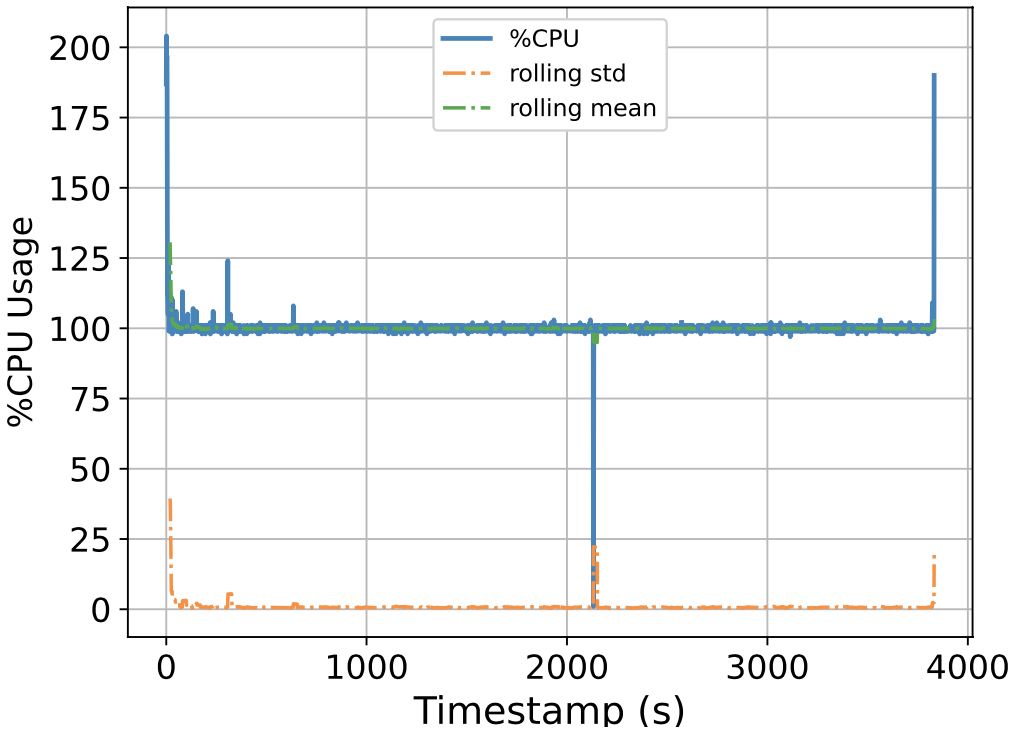
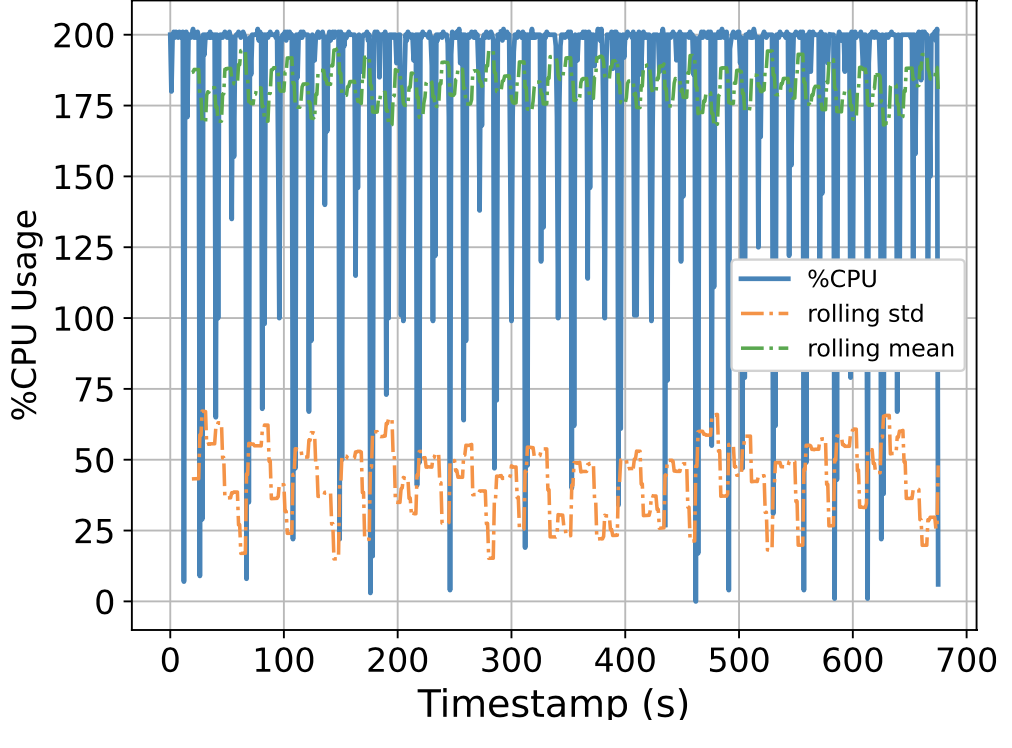
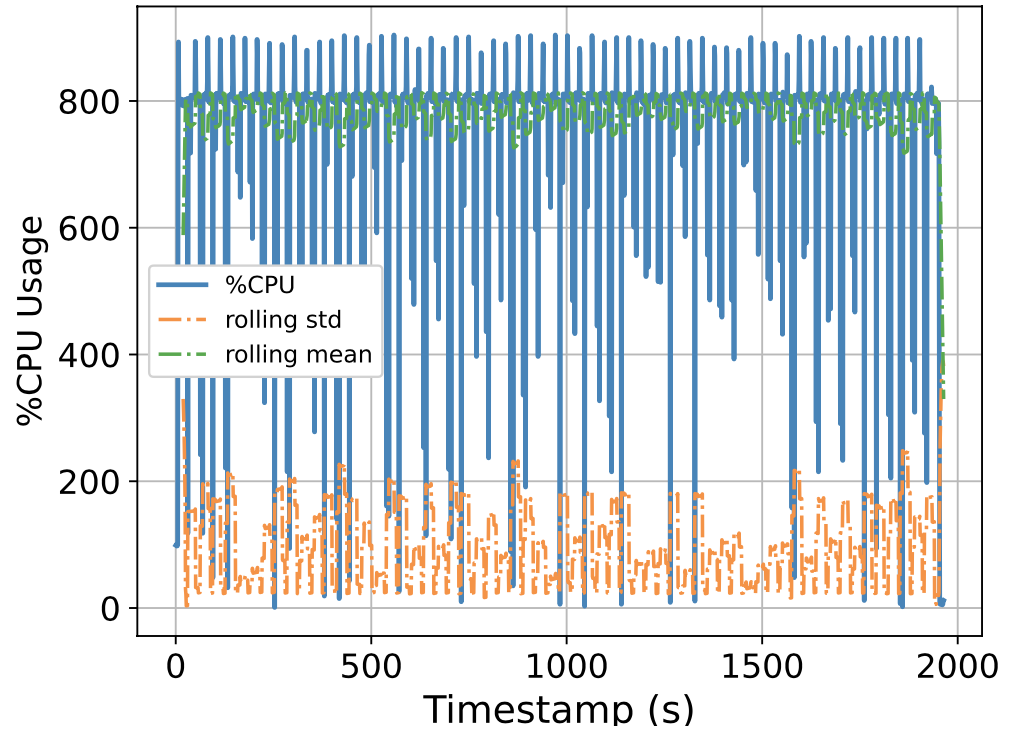
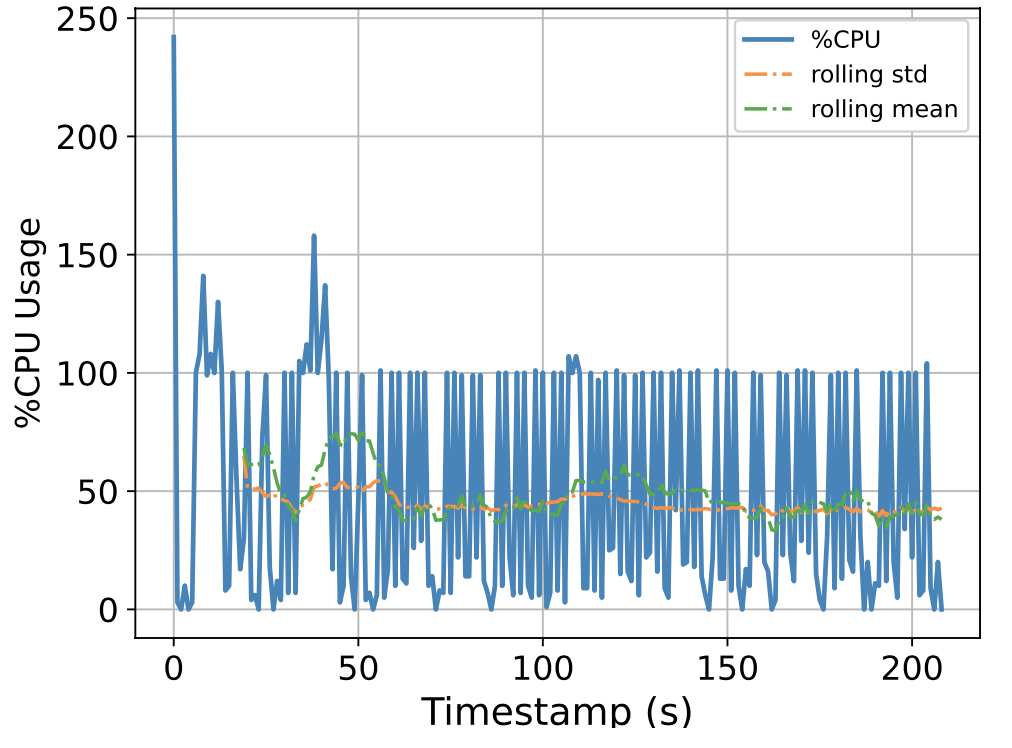
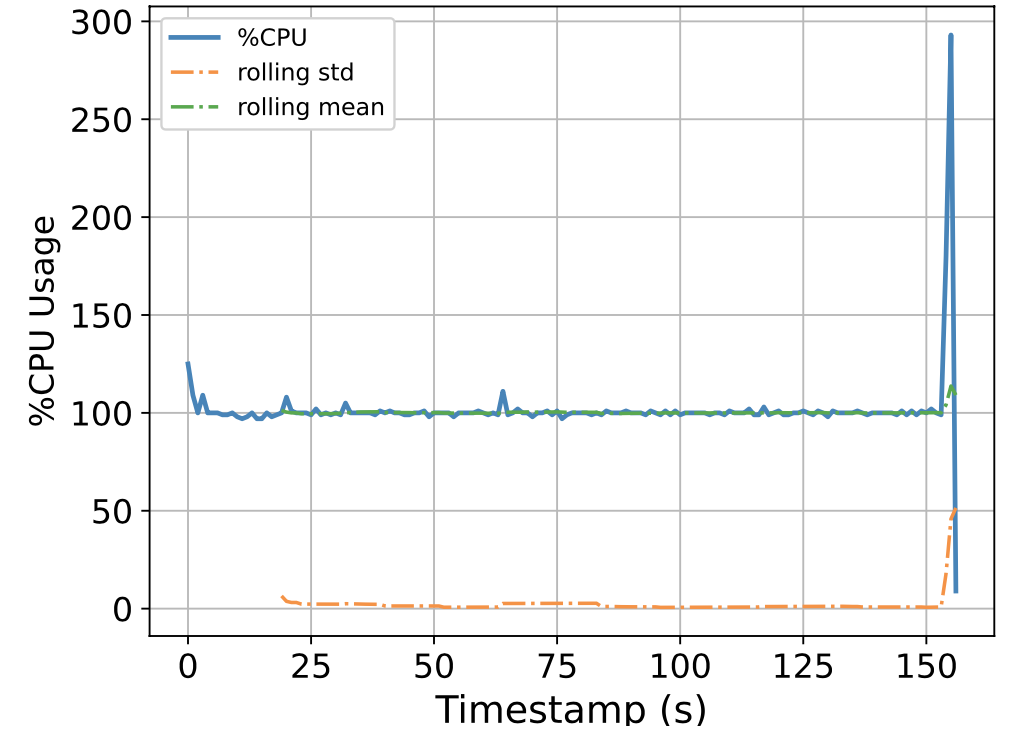
Figure 1. CPU Utilization Patterns. Each subfigure shows the CPU utilization pattern for 9 applications used in a popular genomics workflow. Their dynamicity suggests opportunities to reclaim compute resources at a fine-grained level. x-axis: time since application starts (seconds), y-axis: amount of cores used x 100%.
These initial results provided us with an interesting observation: even for highly parallelizable applications which we thought will completely saturate a machine's CPU resources, their utilization patterns are dynamics (there are recurring up and lows) due to interleaving I/Os operations, and there are resource reclamation opportunities at a fine-grained level. We build upon this observation and devise a resource reclamation technique specifically tailored for these genomics applications. This is one example of how the data generated by our benchmarking tool can guide the design of novel systems support.
Running the Experiment on Chameleon
As we aim to build a benchmarking tool that can characterize performance at scale, it is imperative to also have an infrastructure & enough computational resources to execute the workload at scale. We found Chameleon immensely helpful in this regard. Our tool is designed to run on a cluster, thus we need multiple machines that can easily communicate with each other. We also need a large amount of storage to stage the genomics workflow inputs, which can easily reach tens of GBs per sample. We use the hardware catalog extensively to choose appropriate node types with enough CPU, memory, storage, and I/O bandwidth. We also use Chameleon's CLI to automate a big part of our infrastructure setup.
We are especially fond of Chameleon's bare metal capabilities. It is known that there are performance variations between execution on bare metal vs virtualized environments. By characterizing performance on bare metal and VMs separately, the data we generate becomes richer, and we have solid grounds to claim how certain applications will perform given certain hardwares. Chameleon's object store, with PBs capacity, allows us to store large input files easily. We imagine setting up data storage would be much more difficult without this feature.
We plan to open-source our benchmarking tool once it is ready. Please stay tuned!
User Interview

Tell us a little bit about yourself
I am a 4th year Computer Science PhD student at the University of Chicago. My previous works were on storage systems reliability; this continues to interest me, but I'm currently focused on designing and building novel systems support for large-scale genomics data processing. I think I stumbled upon a subfield with lots of interesting & potentially impactful problems. If possible, I would like to continue a similar line of work, preferably in academia. Outside of work you might find me in museums, orchestras, or simply reading a book in open space.
Most powerful piece of advice for students beginning research or finding a new research project?
Believe in the process! Keep on doing the good habits. You will be surprised by how far you walked.
How do you stay motivated through a long research project?
I often try to imagine how my work would fit in the 'bigger picture'. This gives me the reassurance that what I'm doing is indeed meaningful, although it's probably small, and each day is filled with failed experiments (alas). I also try to cherish the small wins – any improvement, however small, might be a sign that we're in the right direction. Keep pushing!
Are there any researchers you admire? Can you describe why?
There are people whom I think are very bold & visionary in their works. They believe that the field is moving / should move towards a certain direction, they anticipate the challenges, then their works 'ease' the way for the whole community to move forward. I think it's a very admirable way of doing science.
Thank you for sharing your knowledge with the Chameleon community!
Rethinking Memory Management for Multi-Tiered Systems
Exploring Efficient Page Profiling and Migration in Large Heterogeneous Memory
- July 22, 2024 by
- Dong Li
Explore the cutting-edge research of Professor Dong Li from UC Merced as he tackles the challenges of managing multi-tiered memory systems. Learn how his innovative MTM (Multi-Tiered Memory Management) system optimizes page profiling and migration in large heterogeneous memory environments. Discover how Chameleon's unique hardware capabilities enabled this groundbreaking experiment, and gain insights into the future of high-performance computing memory management. This blog offers a glimpse into the complex world of computer memory hierarchies and how researchers are working to make them more efficient and accessible.
Real-time Scheduling for Time-Sensitive Networking: A Systematic Review and Experimental Study
Optimizing Network Performance with Chameleon's Computing Power
- June 25, 2024 by
- Chuanyu Xue
In this study, Chuanyu Xue tackles the complex challenge of optimizing Time-Sensitive Networking (TSN) for real-world applications. Using Chameleon's powerful computing resources, he conducts a comprehensive evaluation of 17 scheduling algorithms across 38,400 problem instances. This research not only sheds light on the strengths and weaknesses of various TSN scheduling methods but also demonstrates how large-scale experimentation can drive advancements in network optimization. Readers will gain insights from Xue's journey, including key findings, implementation challenges, and valuable tips for leveraging Chameleon in their own research.
Optimizing Production ML Inference for Accuracy and Cost Efficiency
Pushing the Boundaries of Cost-Effective ML Inference on Chameleon Testbed
- May 28, 2024 by
- Saeid Ghafouri
In this blog post, we explore groundbreaking research on optimizing production ML inference systems to achieve high accuracy while minimizing costs. A collaboration between researchers from multiple institutions has resulted in the development of three adaptive systems - InfAdapter, IPA, and Sponge - that tackle the accuracy-cost trade-off in complex, real-world ML scenarios. Learn how these solutions, implemented on the Chameleon testbed, are pushing the boundaries of cost-effective ML inference and enabling more accessible and scalable ML deployment.
No comments