Minimizing Out-of-Memory Failures in Genomics Workflow Execution
Reducing Workflow Failures with Chameleon’s Scalable Research Platform
- Dec. 30, 2024 by
- Aaditya Mankar
Delivering high throughput for large-scale genomics data processing is both costly and challenging. Executing genomics workflows requires extensive computing clusters, often resulting in operational costs that reach millions of dollars annually. The challenge arises from the diverse nature of genomics workflows, which encompass a diverse set of applications, each with unique resource demands, utilization patterns, and large input datasets. Workflow execution must be carefully scheduled to avoid failures (e.g., out-of-memory errors) while minimizing costs, such as optimizing the number of machines used. Moreover, these objectives must often be achieved within specific deadline constraints.
While we recognize that this is an instance of a workflow scheduling problem, which is a well-known problem, solving it is particularly challenging due to its domain-specific nature of genomic processing. Simply applying techniques from other domains may be suboptimal due to the difference in workload resource requirements and utilization patterns. Addressing this challenge requires (a) an in-depth understanding of our workload characteristics and also (b) a way to rapidly test our new ideas at a scale that is representative of production deployment.
Research Overview
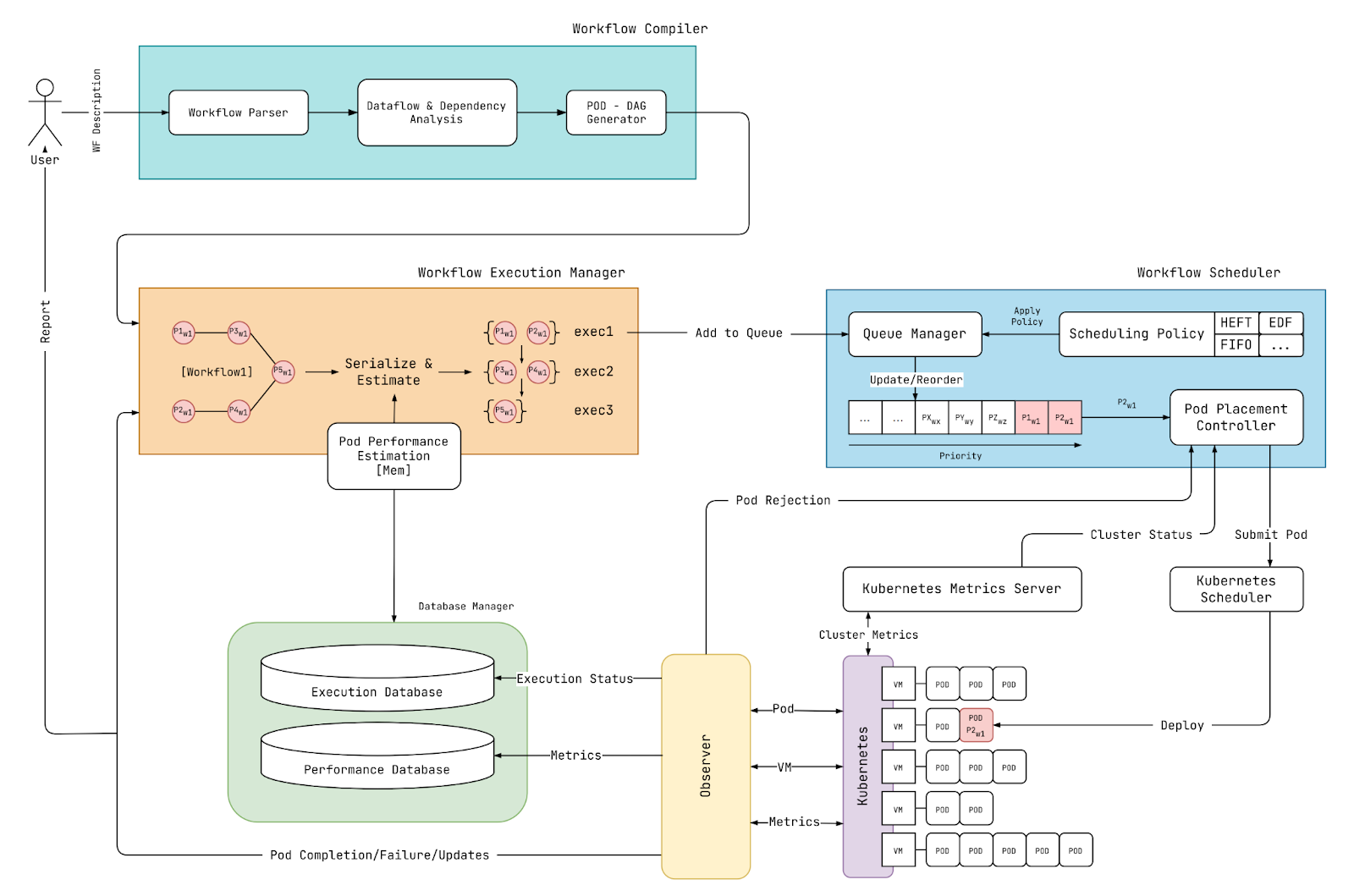
Figure 1. Design of our scalable benchmark tool and scheduler research platform for genomic workflows.
To this end, we are developing the system illustrated in Figure 1, which comprises (1) a scalable benchmarking tool and (2) a scheduler research platform tailored for genomics data processing. Below, we demonstrate its potential benefits by offering insights into minimizing out-of-memory failures during workflow execution.
Failure-Aware Scheduler
Accurately estimating memory and CPU usage during workflow execution is critical to avoiding the drawbacks of resource misallocation. Overallocation leads to resource waste, as idle resources allocated to one job remain unavailable for others. Underallocation, however, can result in performance degradation. While CPU is a compressible resource, making its misallocation less critical, underallocating memory can lead to out-of-memory failures, halting genomic workflow executions and wasting all time and resources allocated and consumed up to the point of failure.
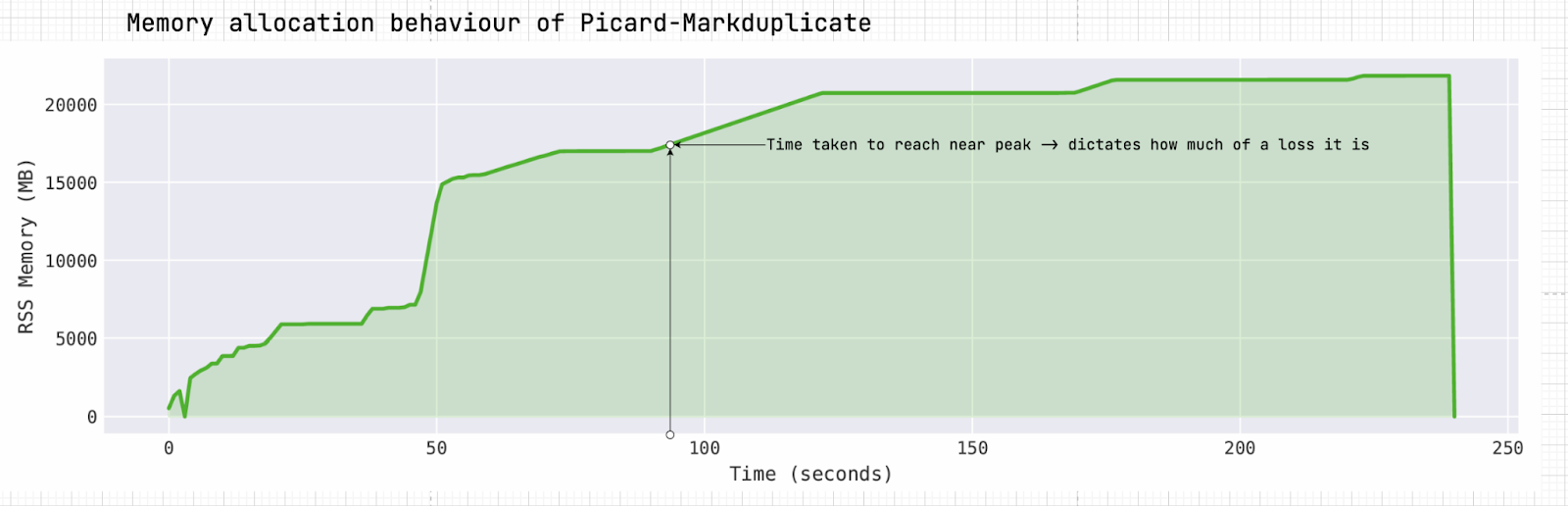
Figure 2. Memory utilization overtime of Picard-MarkDuplicate.
A common approach to memory allocation relies on peak memory usage. However, our workload characterization reveals significant variability in the memory usage of several genomic applications. For example, Picard-MarkDuplicate, a widely used genomic tool for identifying and marking duplicate reads in sequencing data, exhibits this variability as shown in Figure 2. The heatmap in Figure 3 illustrates how peak memory usage fluctuates with input size. For inputs ranging from 100 MB to 1 GB, peak memory usage varies significantly, ranging from 13 GB to 22 GB. This variability forces a choice between overallocating, which wastes resources, or encountering repeated failures while attempting to determine an appropriate “average” value.
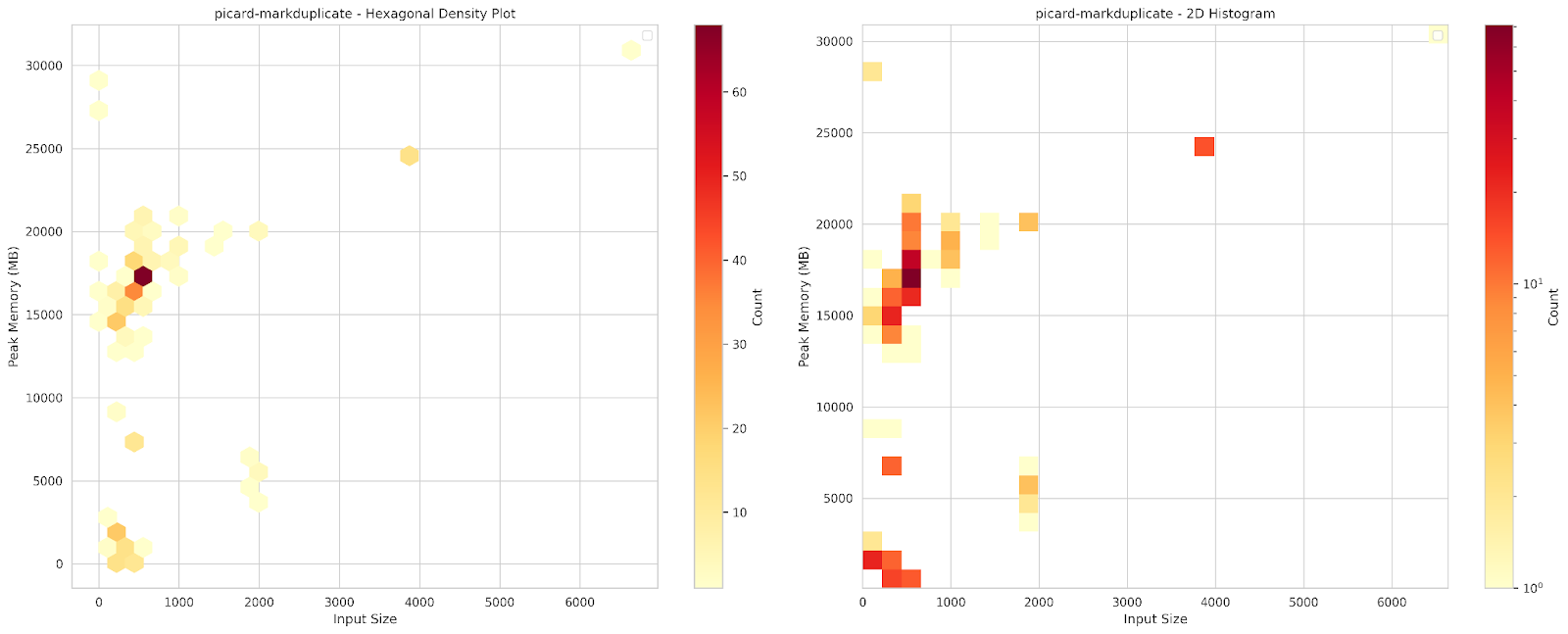
Figure 3. Peak memory of Picard-MarkDuplicate across various input sizes.
This variability is where our Failure-Aware Scheduler comes into play. Its design is built on two key principles. First, it accounts for variability by leveraging resource utilization traces from the benchmarking tool to analyze memory usage patterns. The scheduler considers how genomic applications utilize memory and dynamically adjust resource allocations to minimize both resource waste and out-of-memory failures.
Moreover, the scheduler incorporates applications' runtime behavior into its decision-making process. For applications with high variability and a fast climb to peak memory, allowing a few failed attempts is often acceptable. The resource wastage from each failure is minimal, making this approach more efficient in terms of time and resources. Conversely, for applications with high variability but a slower climb to peak memory, failures become more costly, leading the scheduler to prioritize overallocation.
By considering both memory variance and application behavior, the scheduler aims to predict an effective resource allocation with an acceptable rate of failures. While genomic workloads include some inherent unpredictability, we further enable resource optimization by leveraging predictable patterns wherever possible, ensuring the scheduler remains both practical and efficient.
This work is part of a broader effort to address the challenge of cluster sizing under deadline and cost constraints. Beyond the Failure-Aware Scheduler, we utilize benchmarking results on Chameleon servers to develop models for predicting average CPU usage and execution time. Additionally, we plan to explore portfolio scheduling (as illustrated in Figure 4), which will provide users with a comprehensive performance comparison of various policies prior to execution, allowing them to select those that best align with their preferred metrics (e.g., cost versus deadline trade-offs).
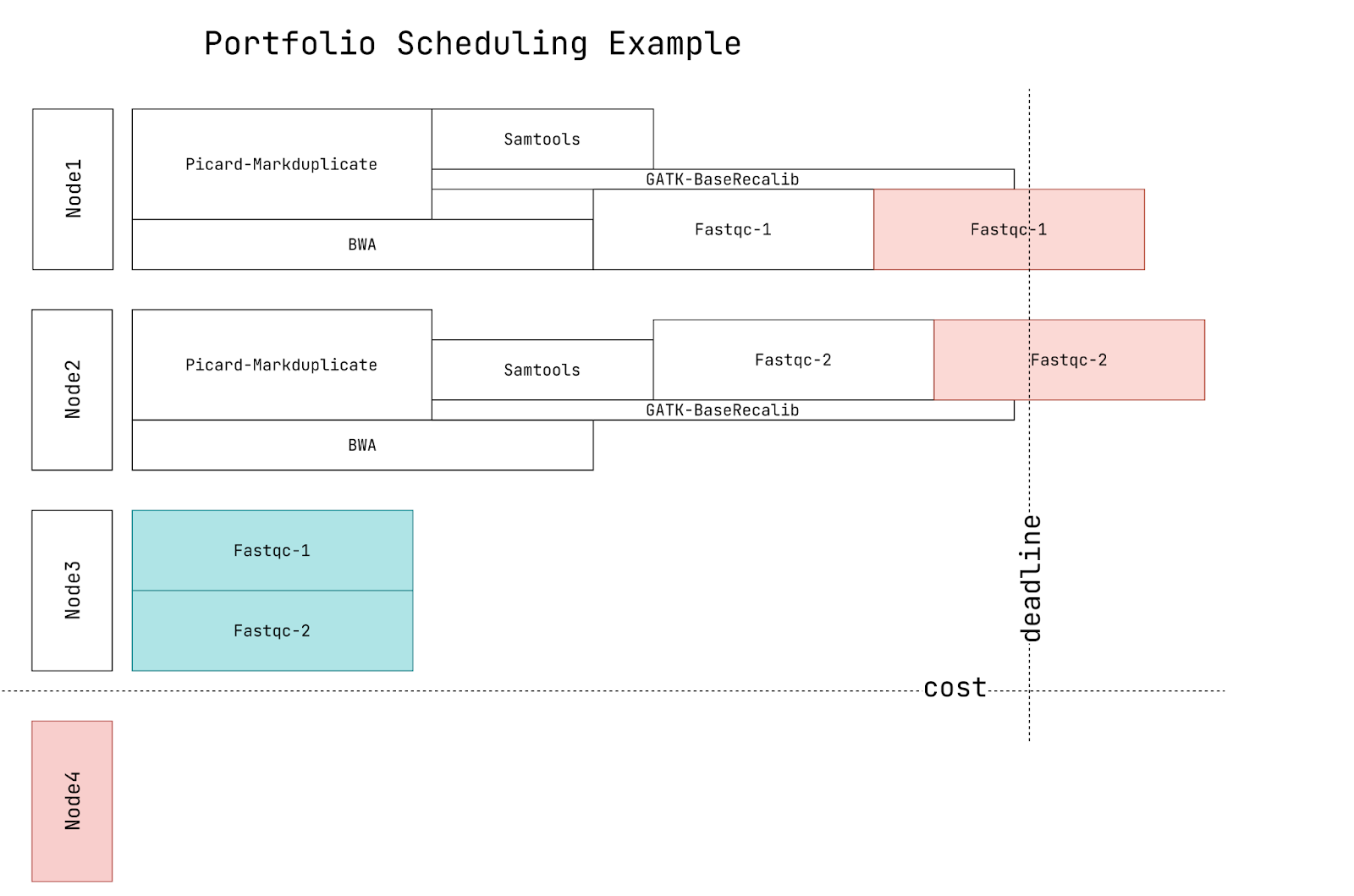
Figure 4. Example of cost-deadline tradeoff in portofolio scheduling.
Running the Experiment on Chameleon
Chameleon makes it easy and fast to provision virtual machines for scientific experiments, even in large numbers. It is user-friendly, offering a variety of pre-configured images and a straightforward setup process for SSH access.
A key benefit for our research is its networking capabilities. For example, Kubernetes requires a complex network setup, and Chameleon's prebuilt configuration significantly reduces the time needed for this process. This enables users to quickly add or remove machines, similar to the experience with commercial cloud providers. Moreover, the project requires autoscaling capability, and in Chameleon, it is straightforward to create a script that can efficiently acquire or release machines as needed, making resource management flexible and effective.
User Interview

Tell us a little bit about yourself
I’m currently pursuing my Master’s degree at the University of Georgia, where I’m in my second year. My studies are centered around Operating Systems, Distributed Systems, and Cloud Computing—fields that fascinate me because of their complexity and the real-world impact they can have on technology. I’m particularly passionate about optimizing large-scale systems to enhance efficiency, scalability, and resilience, which I believe are key to driving the future of computing.
Looking ahead, I’m eager to work as a research engineer, applying my knowledge and skills to solve challenging real-world problems in these areas. My goal is to contribute to meaningful technological advancements while continuously learning and growing in my field.
Most powerful piece of advice for students beginning research or finding a new research project?
"The best way to predict the future is to invent it." -- Alan Kay. When I think about starting a new project or overcoming a hurdle, this quote reminds me that if I want something to change, I’m the one who has to make it happen. It’s a reminder that I don’t need to wait for answers to appear. Research is about exploring the unknown, asking bold questions, and creating something new. Embracing curiosity and being willing to venture into uncharted territory are essential.
How do you stay motivated through a long research project?
I stay motivated during long research projects because I’ve always loved learning and the thrill of discovering something new. There’s always something to uncover, whether it’s a tiny detail or a breakthrough idea. Even when I get stuck—which happens often in research—it feels like being in an open sandbox. If one approach doesn’t work, you can try another. Sometimes, stepping back and trying a different perspective is all it takes to view the problem in a new way. I also make sure to take silent walks; they give my mind a chance to catch up, sort through the noise, and often lead to unexpected breakthroughs.
Thank you for sharing your knowledge with the Chameleon community!
Towards Characterizing Genomics Workload Performance at Scale
Leveraging Chameleon's Bare Metal Resources to Benchmark Genomics Workflows
- Aug. 26, 2024 by
- Martin Putra
Martin Putra, a 4th year PhD student at the University of Chicago, shares how he used Chameleon to build and test a scalable benchmarking tool for genomics workflows, uncovering insights that could lead to more efficient resource management for these computationally intensive tasks.
No comments